The Ding Lab is using cutting edge microscopy techniques to visualize tumor architecture at unprecedented detail.
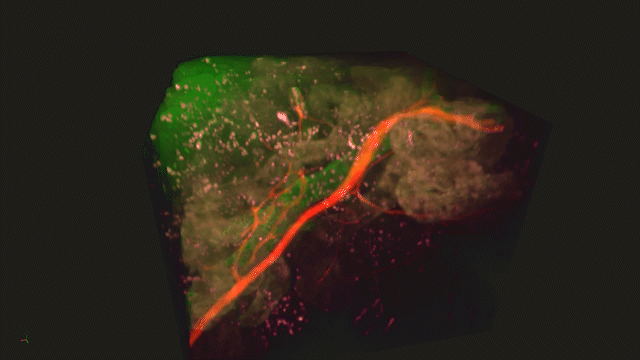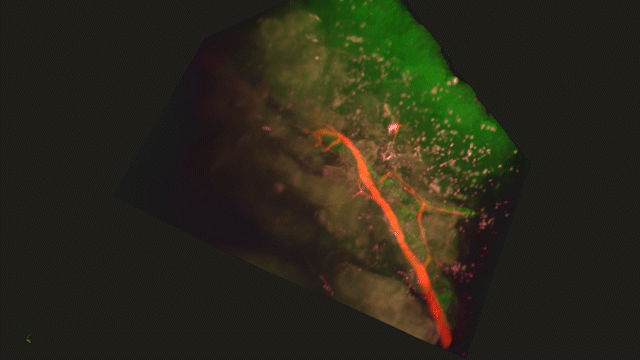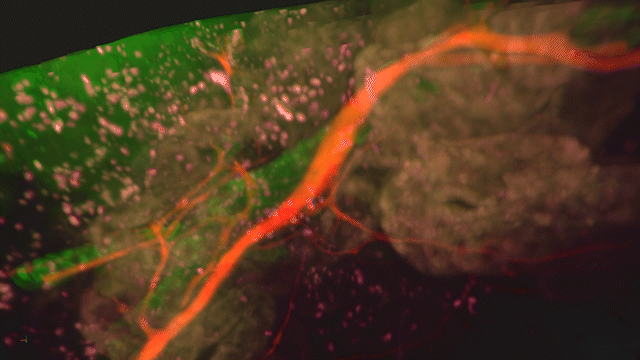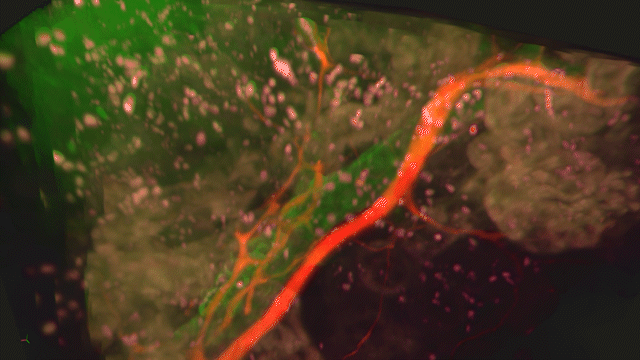
Lightsheet Microscopy
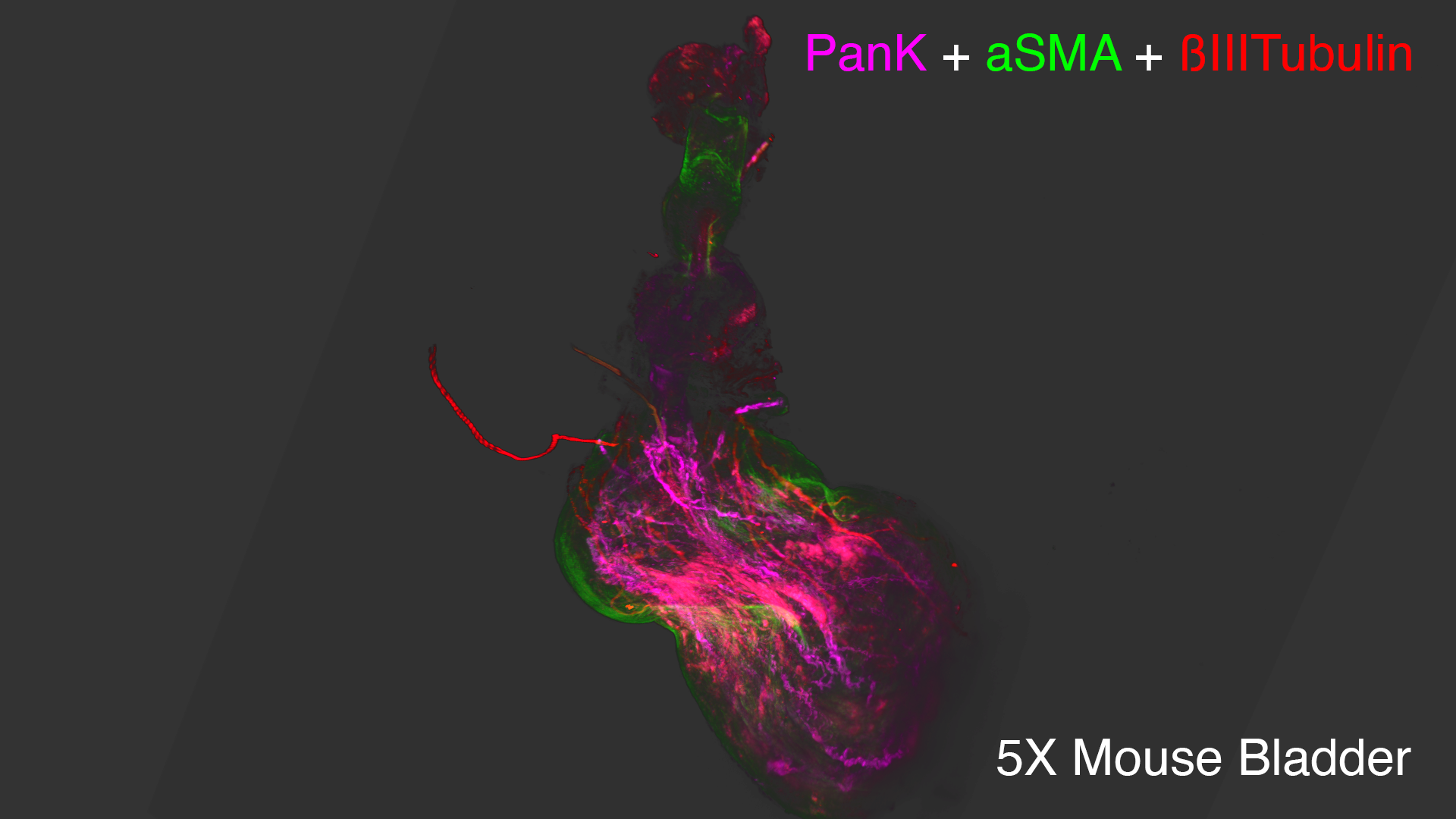
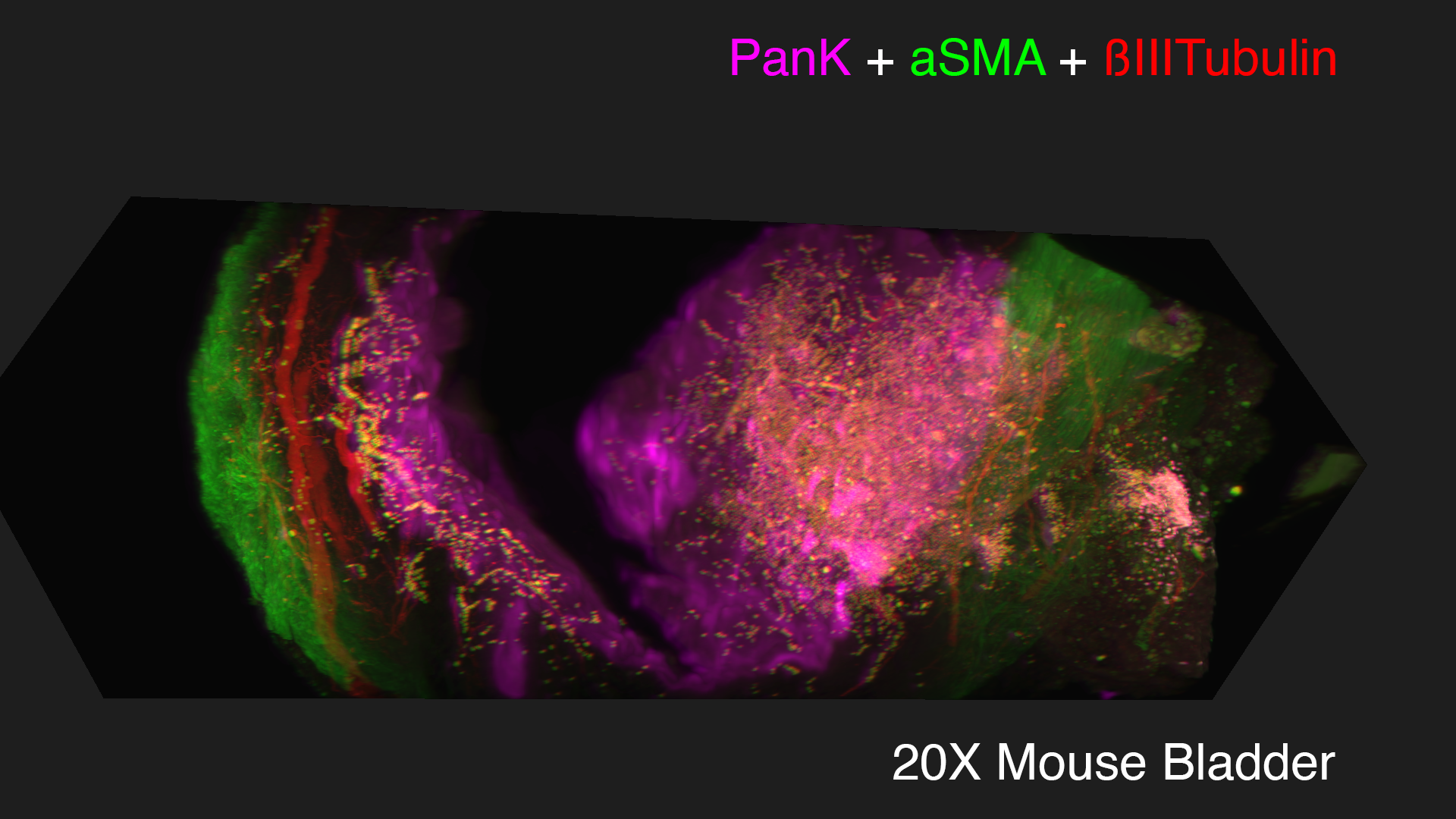
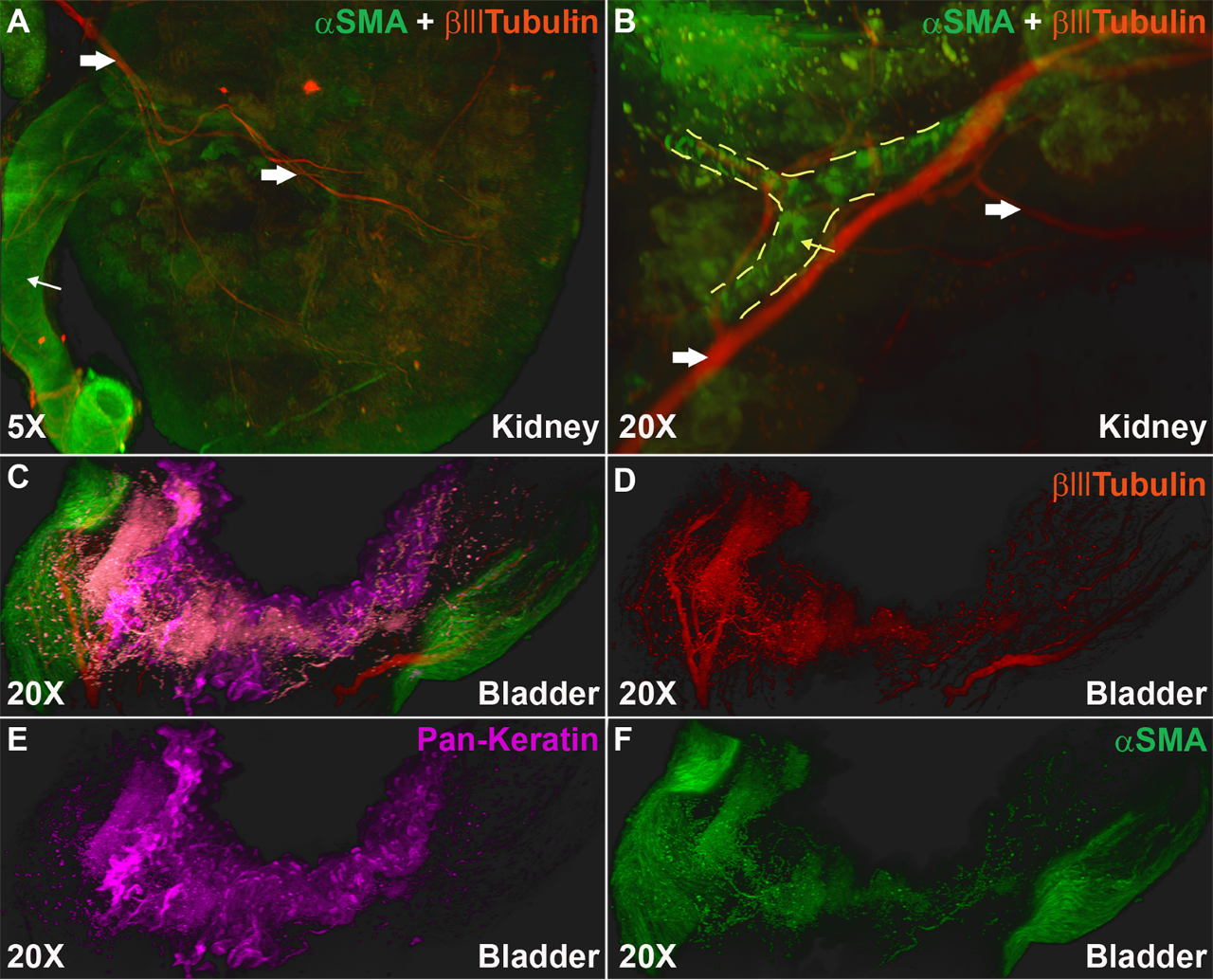
Lightsheet microscopy uses novel techniques to optically clear, or make transparent, various types of tissue. Subsequent staining and three-dimensional imaging of the tissue provides unprecedented insight into the structure within the tissue.
We use Lightsheet Microscopy (LSM) for three-dimensional (3D) multiplexed imaging of large volumes (up to 5 x 10 x 10 mm) of optically-cleared fluorescently labeled samples. LSM offers enhanced speed, resolution, multiplexity and depth with reduced photobleaching. Using LSM with cleared tumor samples, we can reveal 3D architecture of previously unknown features of human solid tumor tissue, including the complex spatial interactions between tumor, immune and stromal cells with vascular and lymphatic vessels, and the extracellular matrix.
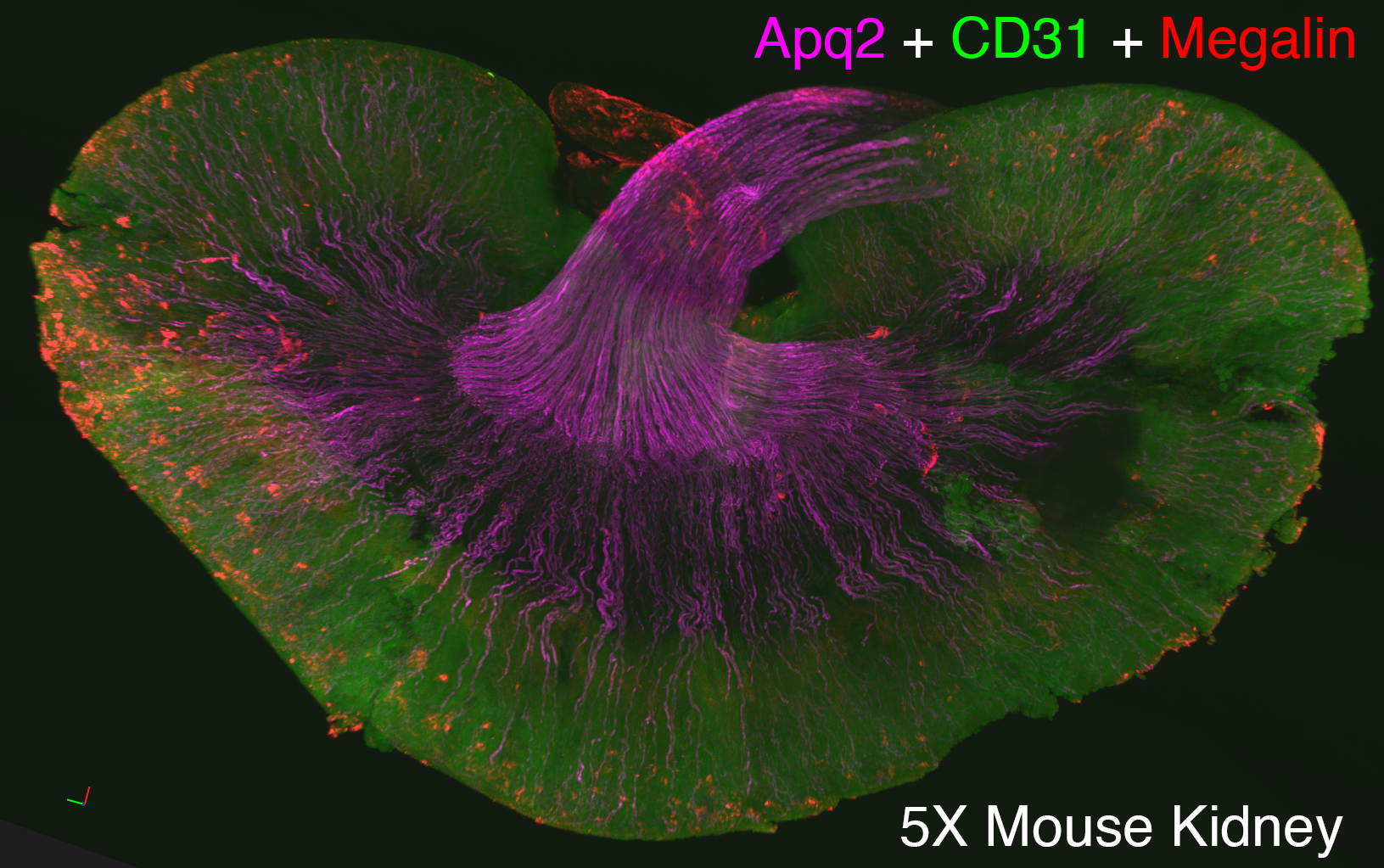
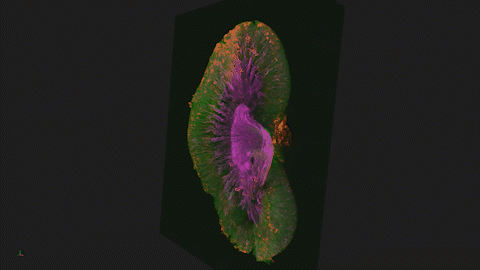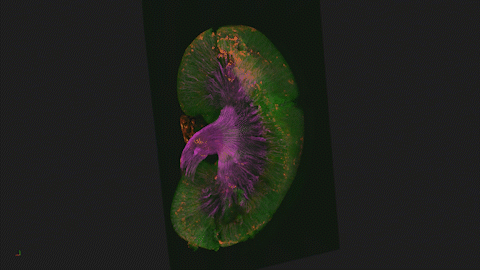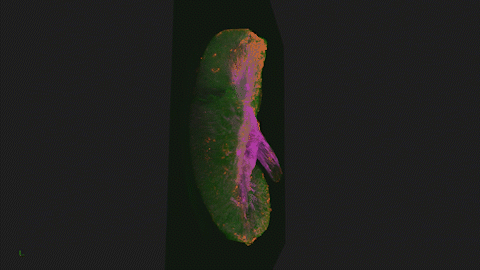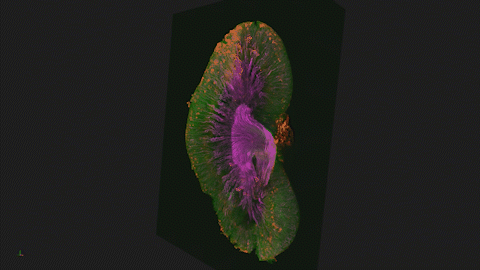
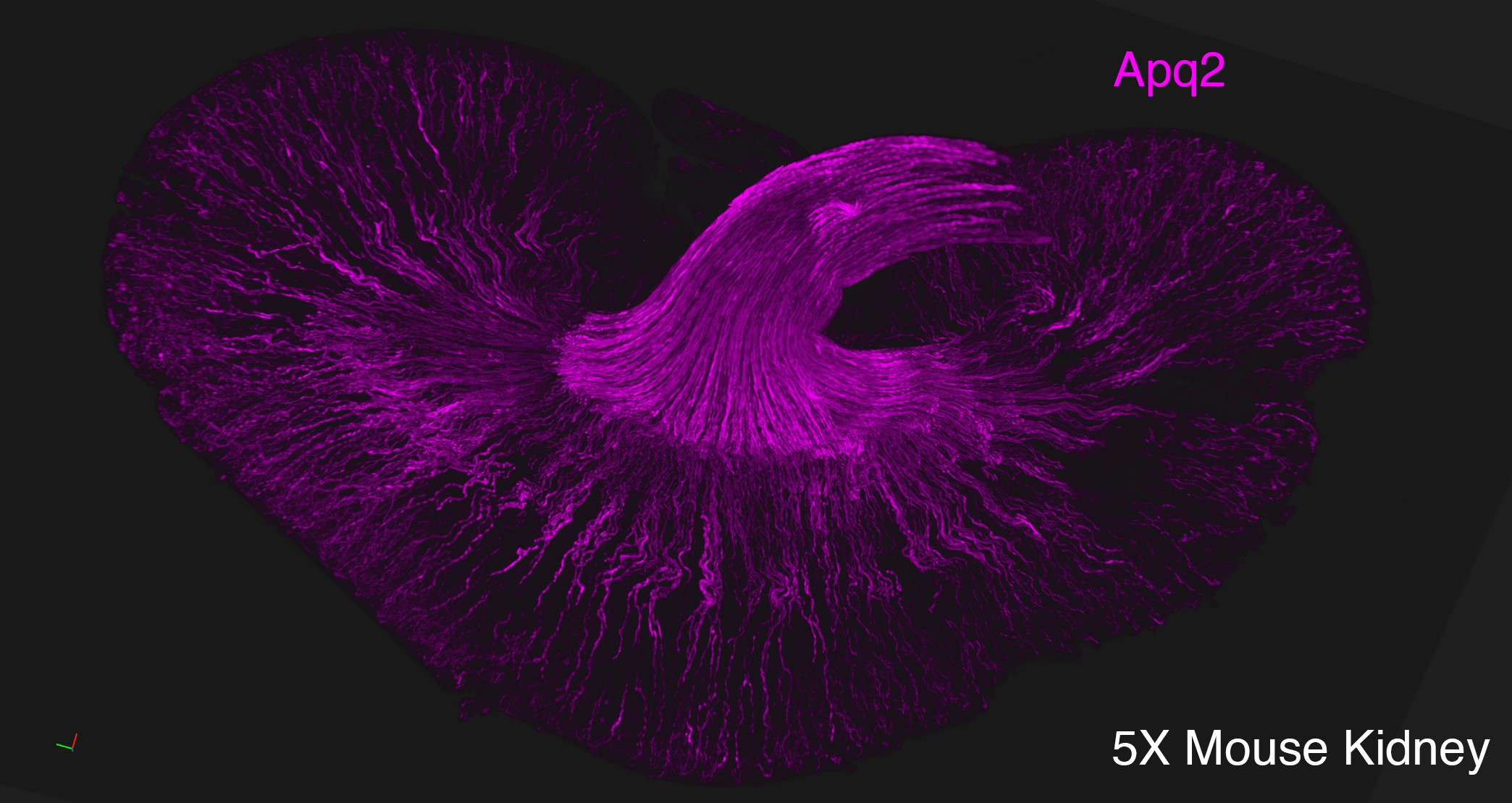
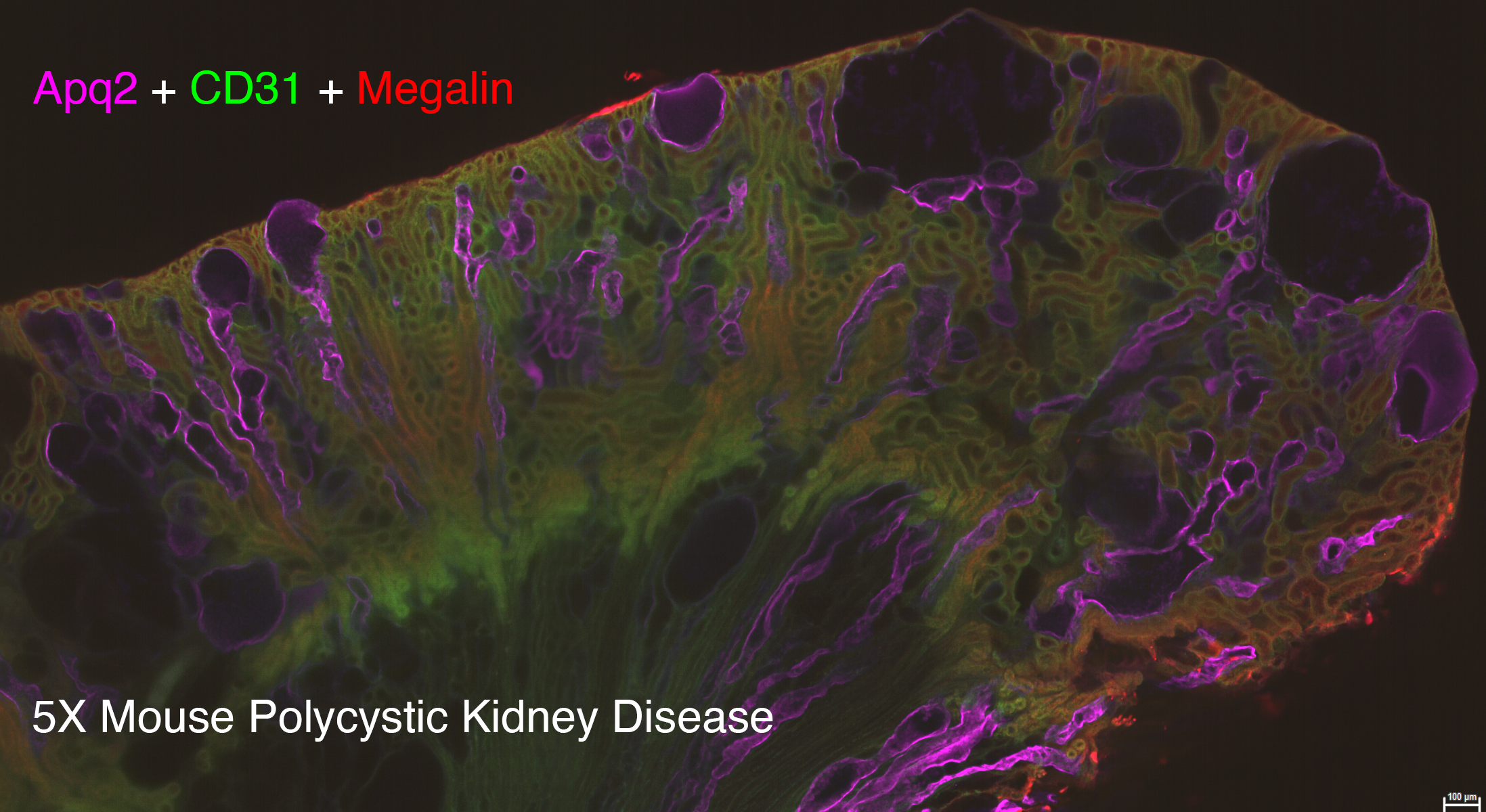
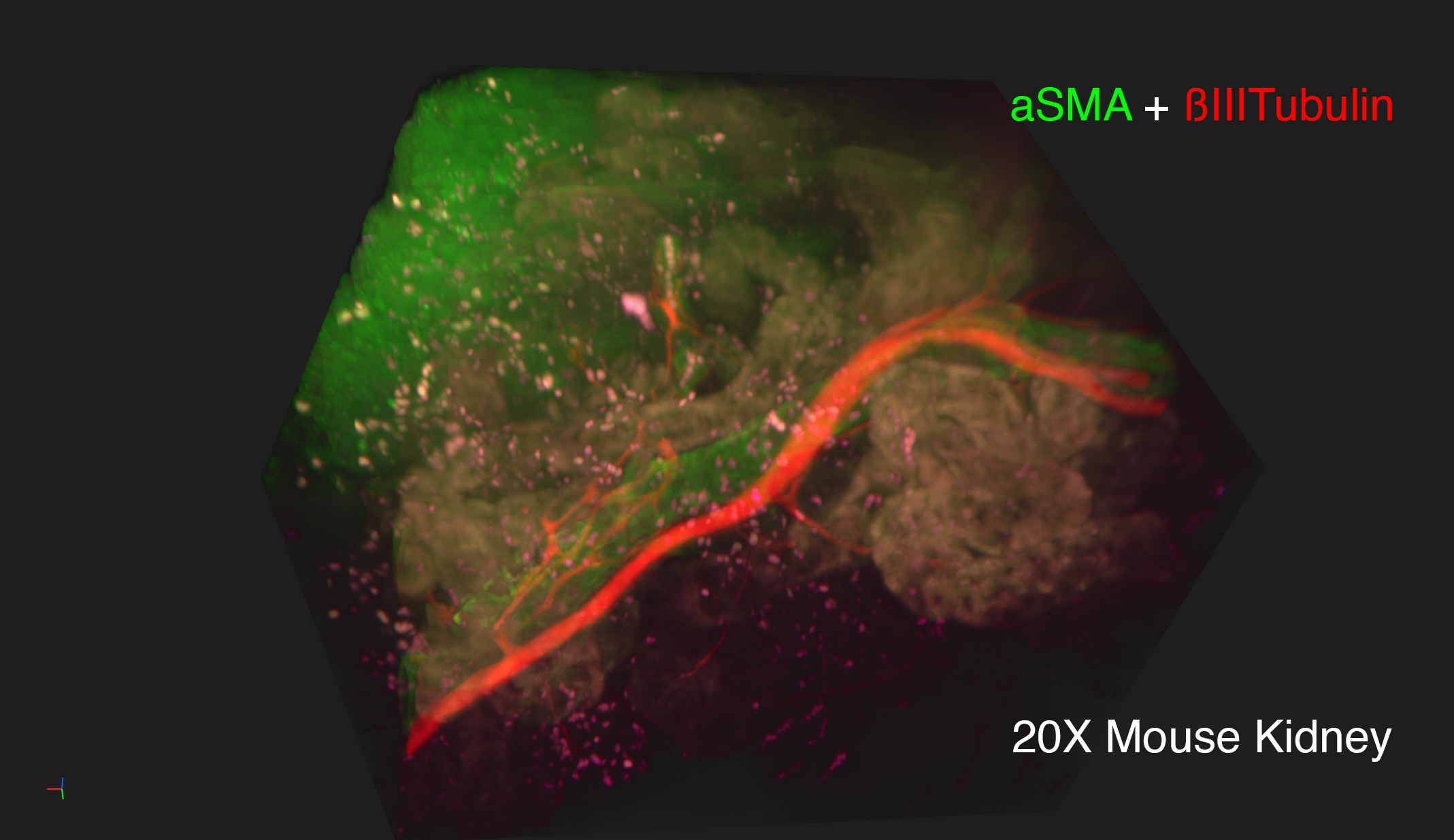
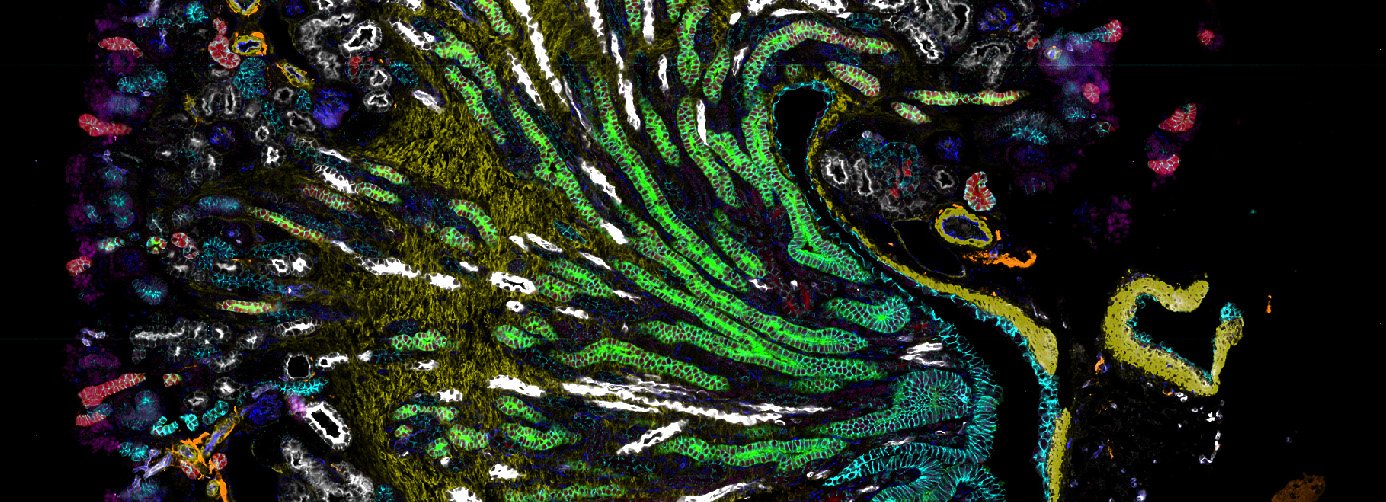
Codex
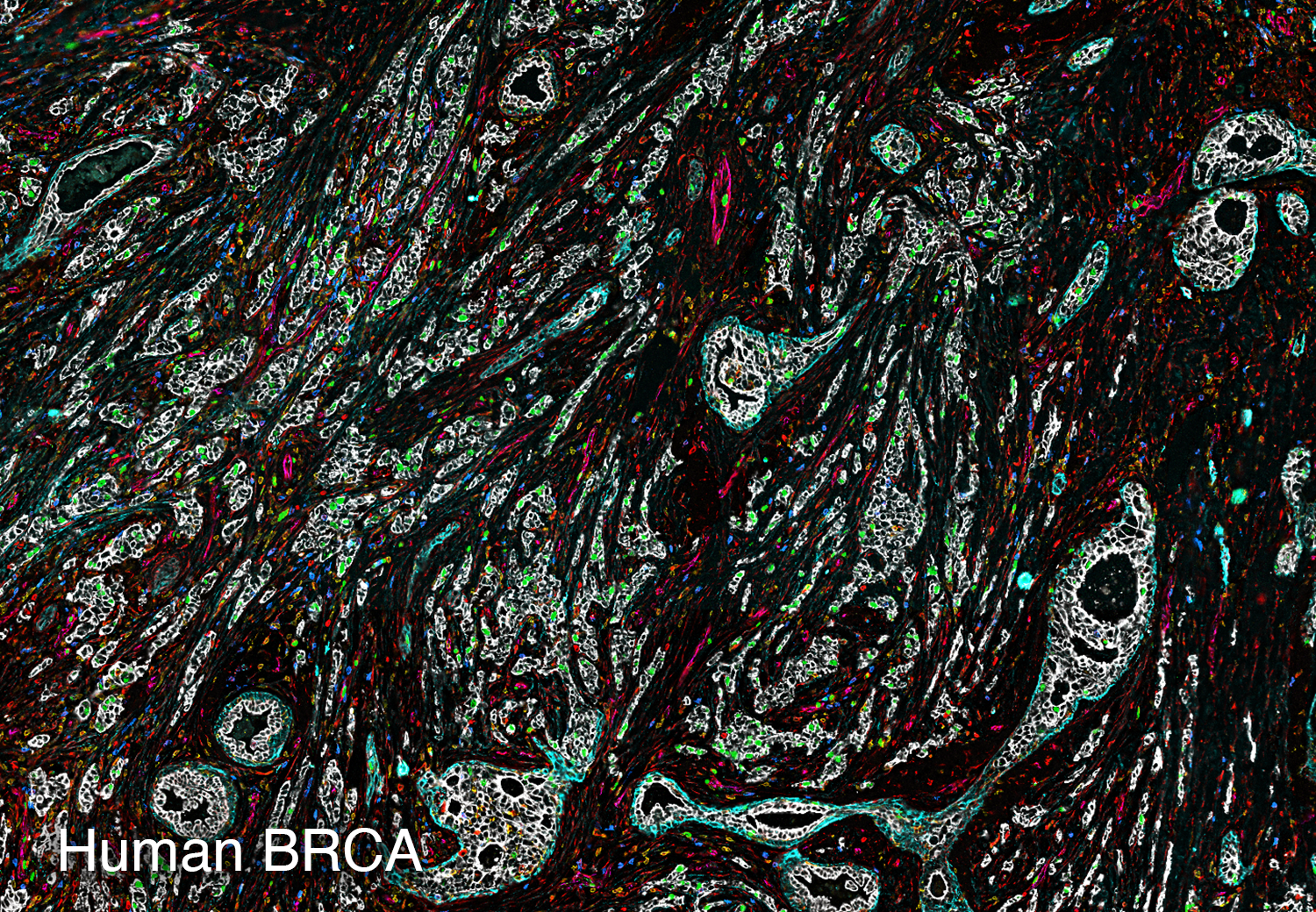
To study the heterogeneity and complexity of different kinds of disease, we apply 30+ antibodies on a single tissue section and use an automated imaging system to produce whole-tissue immunofluorescent images at single cell resolution.
The images below represent tumors from a variety of cancer types. Colors correspond to various proteins expressed by cells within the tumors, and illustrate the complex and diverse tumor microenvironment.
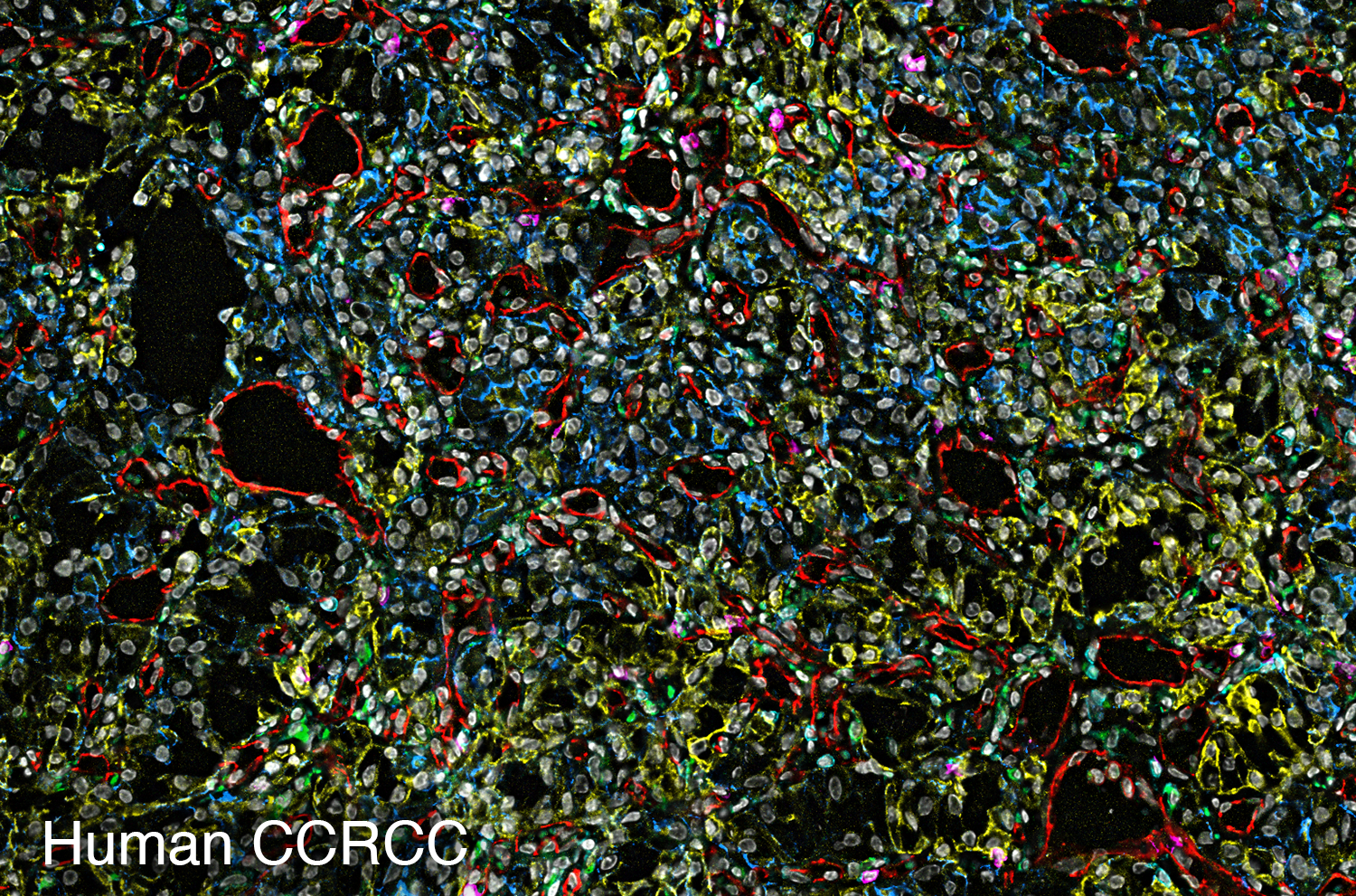
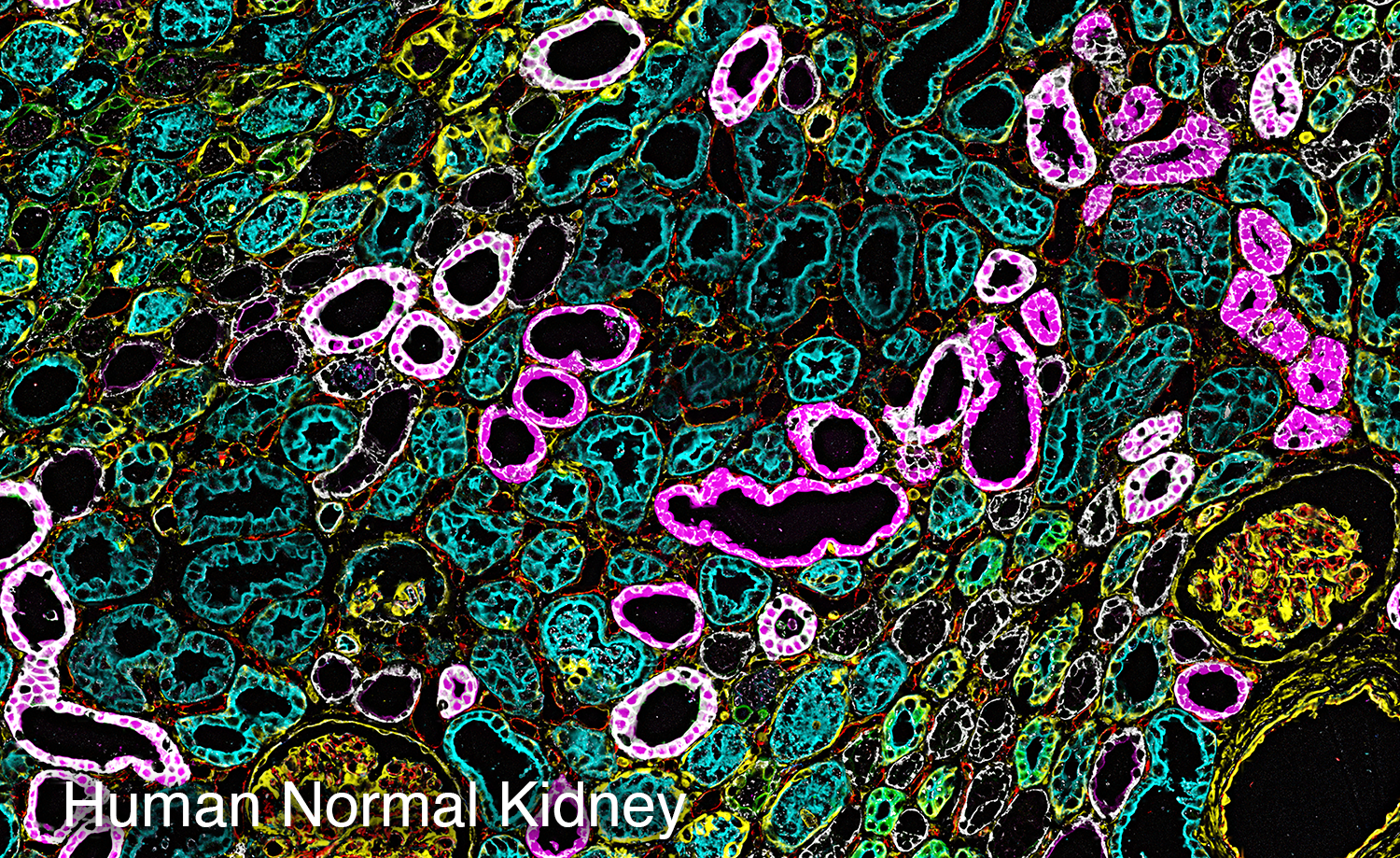
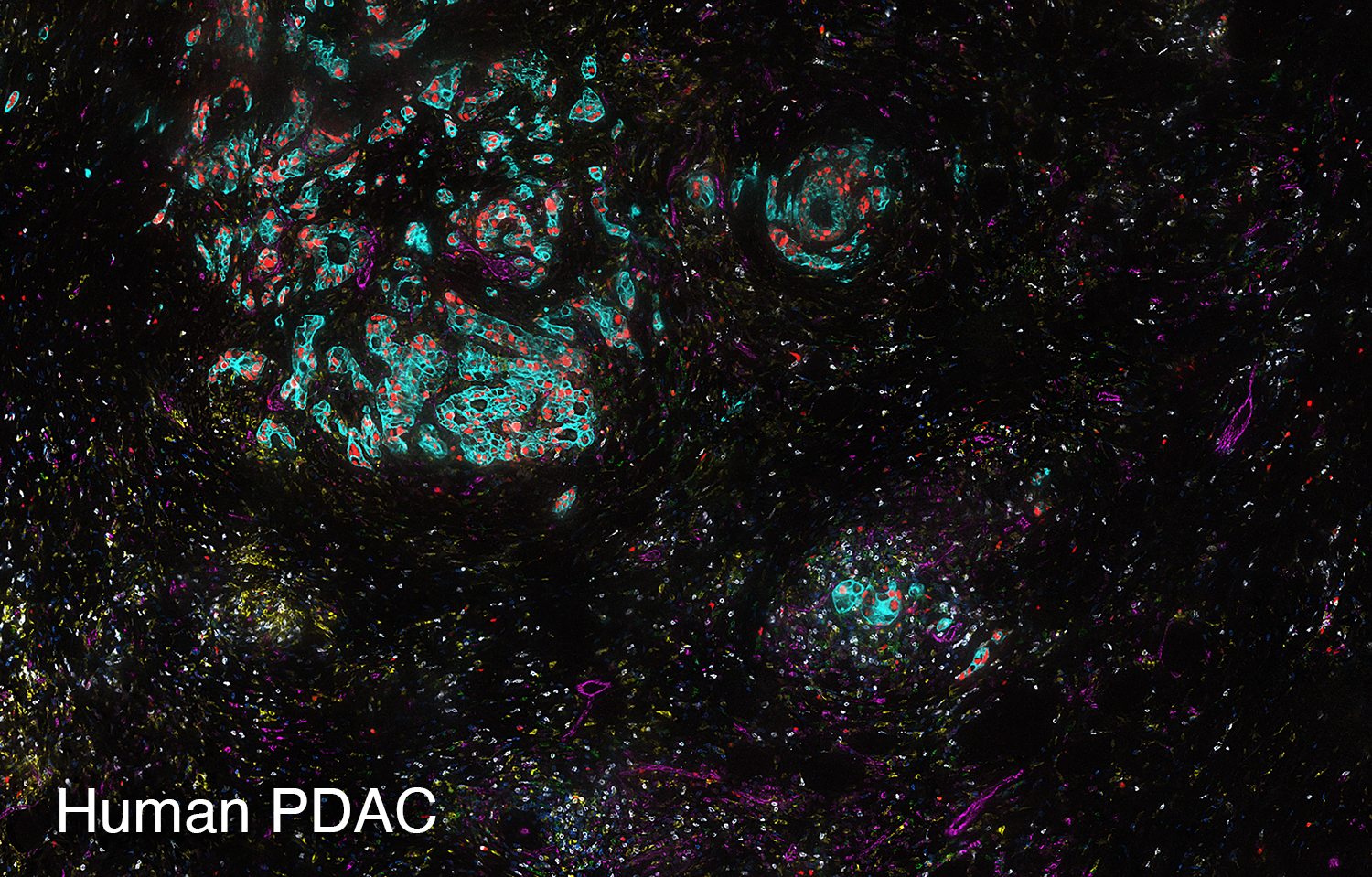
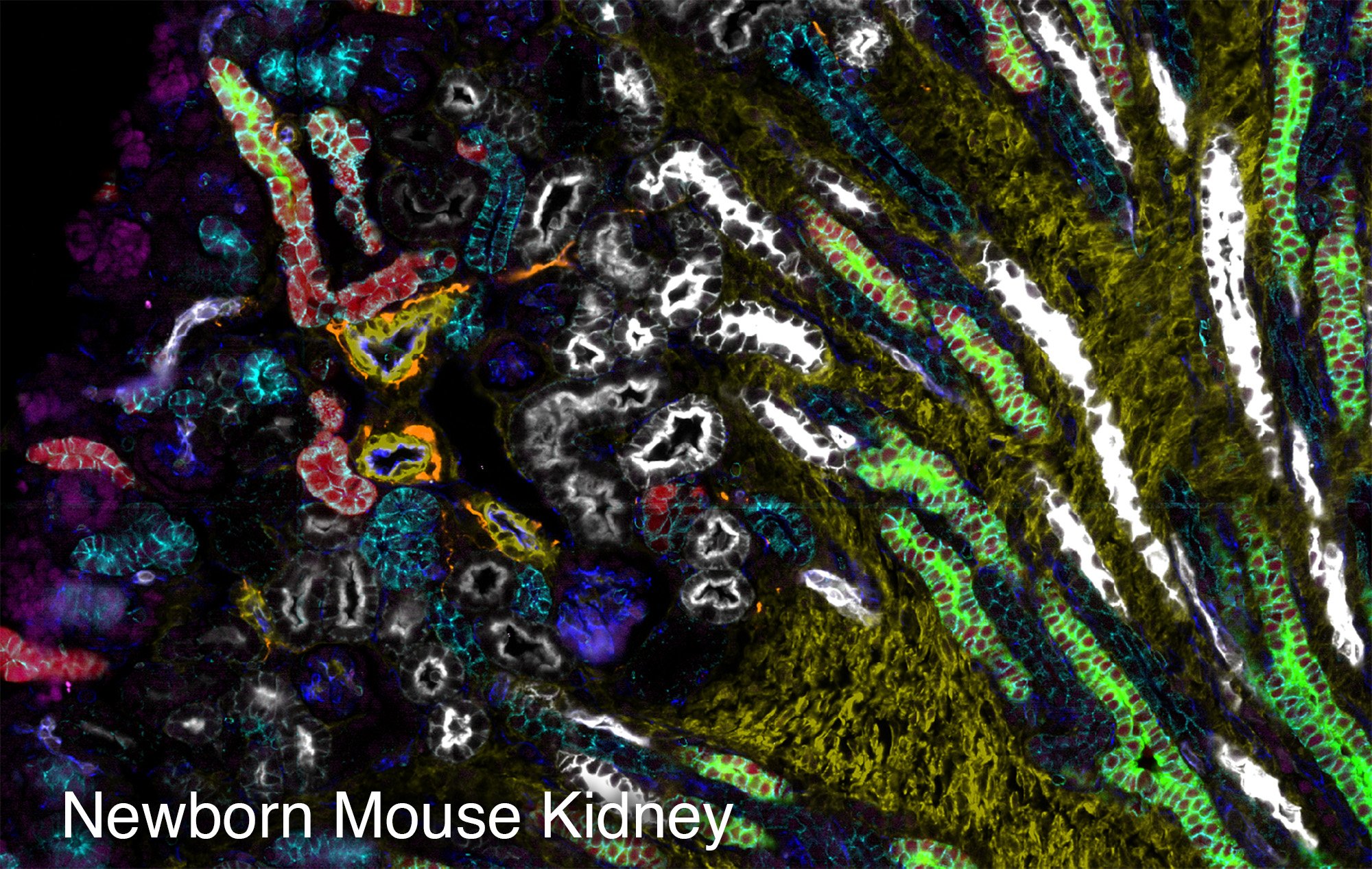
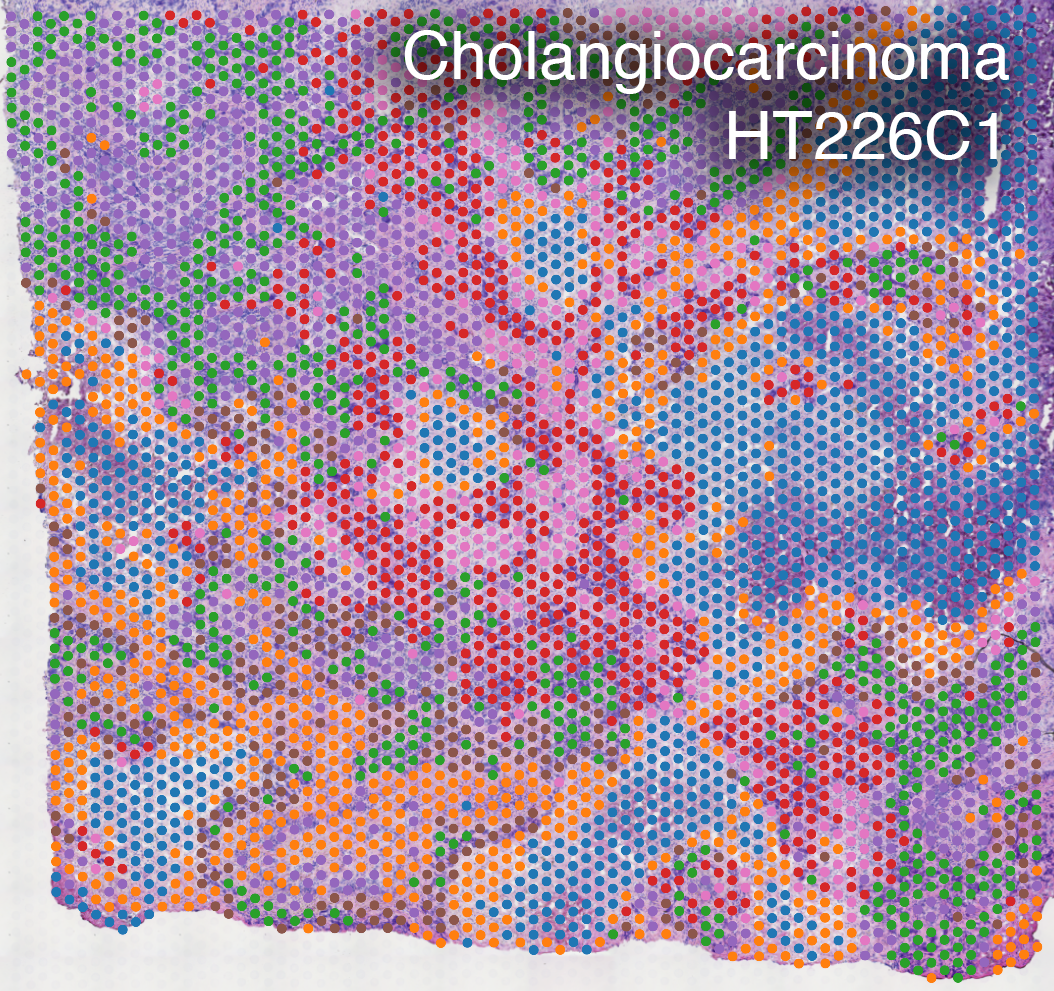
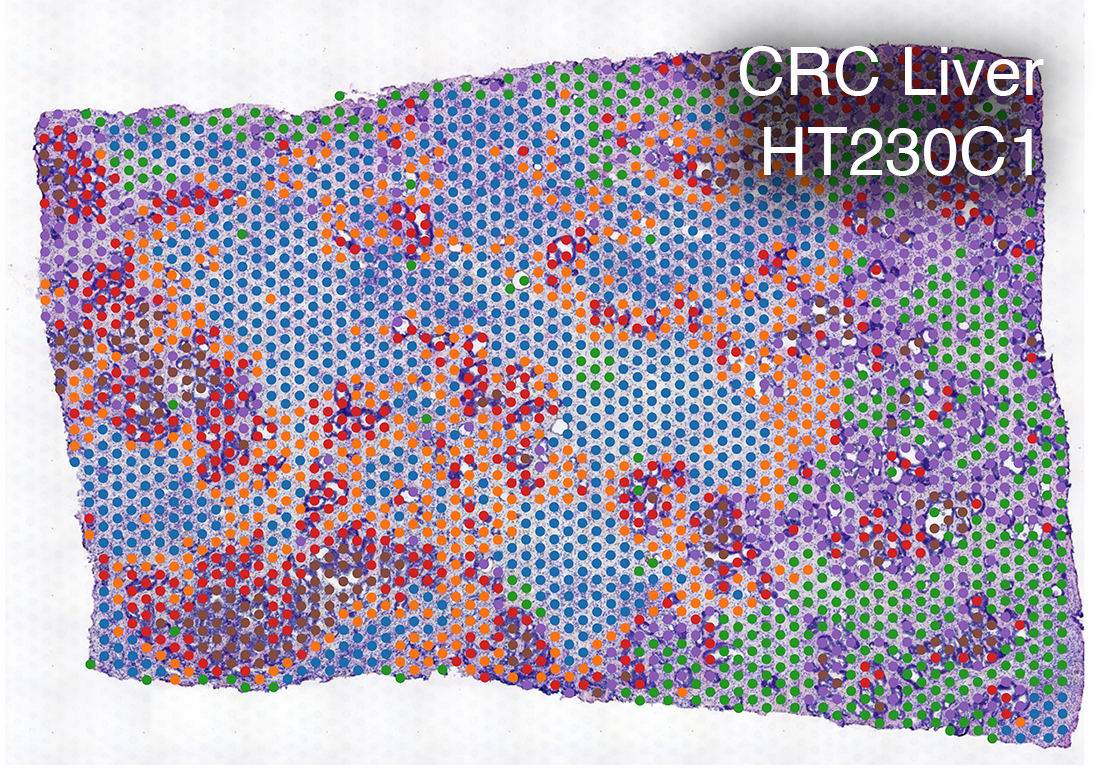
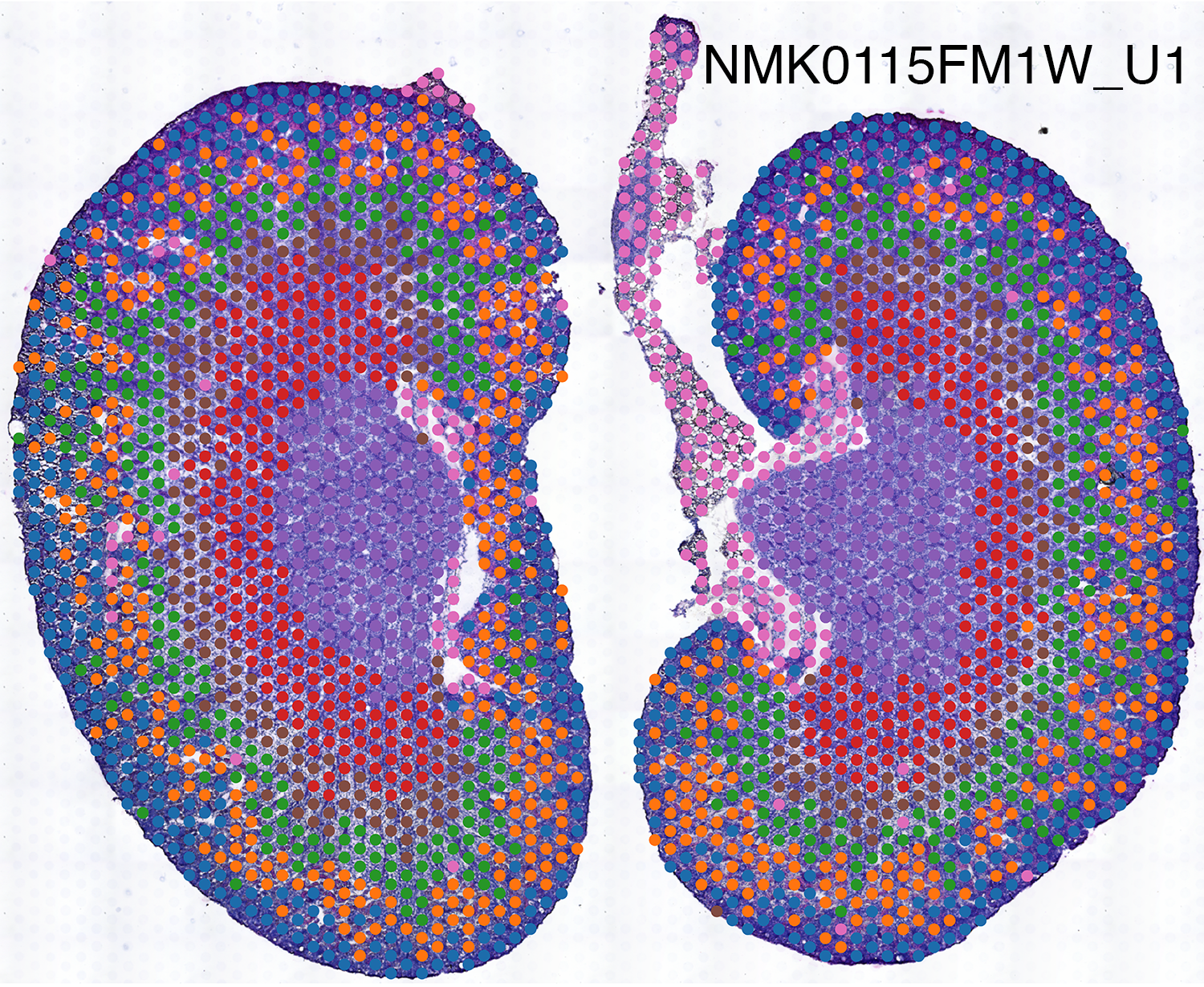
Spatial Transcriptomics
We use spatially-resolved transcriptome to study and discover cellular and molecular interactions such as tumor and immune cells in close proximity.
Spatial transcriptomics combines microscopy imaging with spatially-resolved RNA sequencing. In the images below, histopathology images are overlaid with a grid of dots. RNA in the tissue at each dot location is sequenced. The color of each dot corresponds to the types of cells present there, as indicated by their RNA expression patterns.